Download the paper here
We present a denoising diffusion probabilistic model for protein structure and sequence which conditions on compact protein topology priors to generate proteins.
Abstract
Proteins are macromolecules that mediate a significant fraction of the cellular processes that underlie life. An important task in bioengineering is designing proteins with specific 3D structures and chemical properties which enable targeted functions. To this end, we introduce a generative model of both protein structure and sequence that can operate at significantly larger scales than previous molecular generative modeling approaches. The model is learned entirely from experimental data and conditions its generation on a compact specification of protein topology to produce a full-atom backbone configuration as well as sequence and side-chain predictions. We demonstrate the quality of the model via qualitative and quantitative analysis of its samples.
Protein Backbone Generation
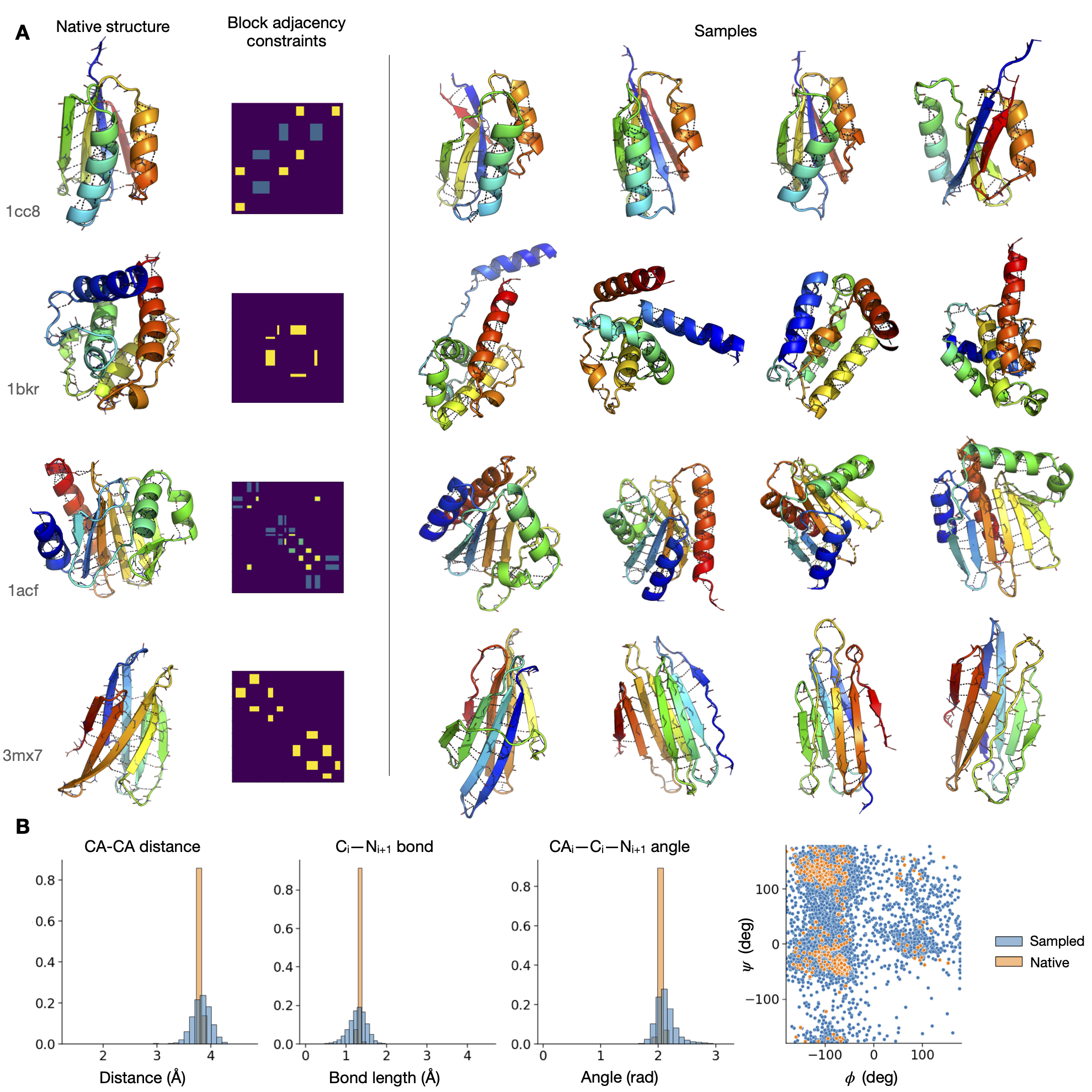
From-scratch generation of proteins by the model. The model conditions only on secondary structure and coarse adjacency constraints.
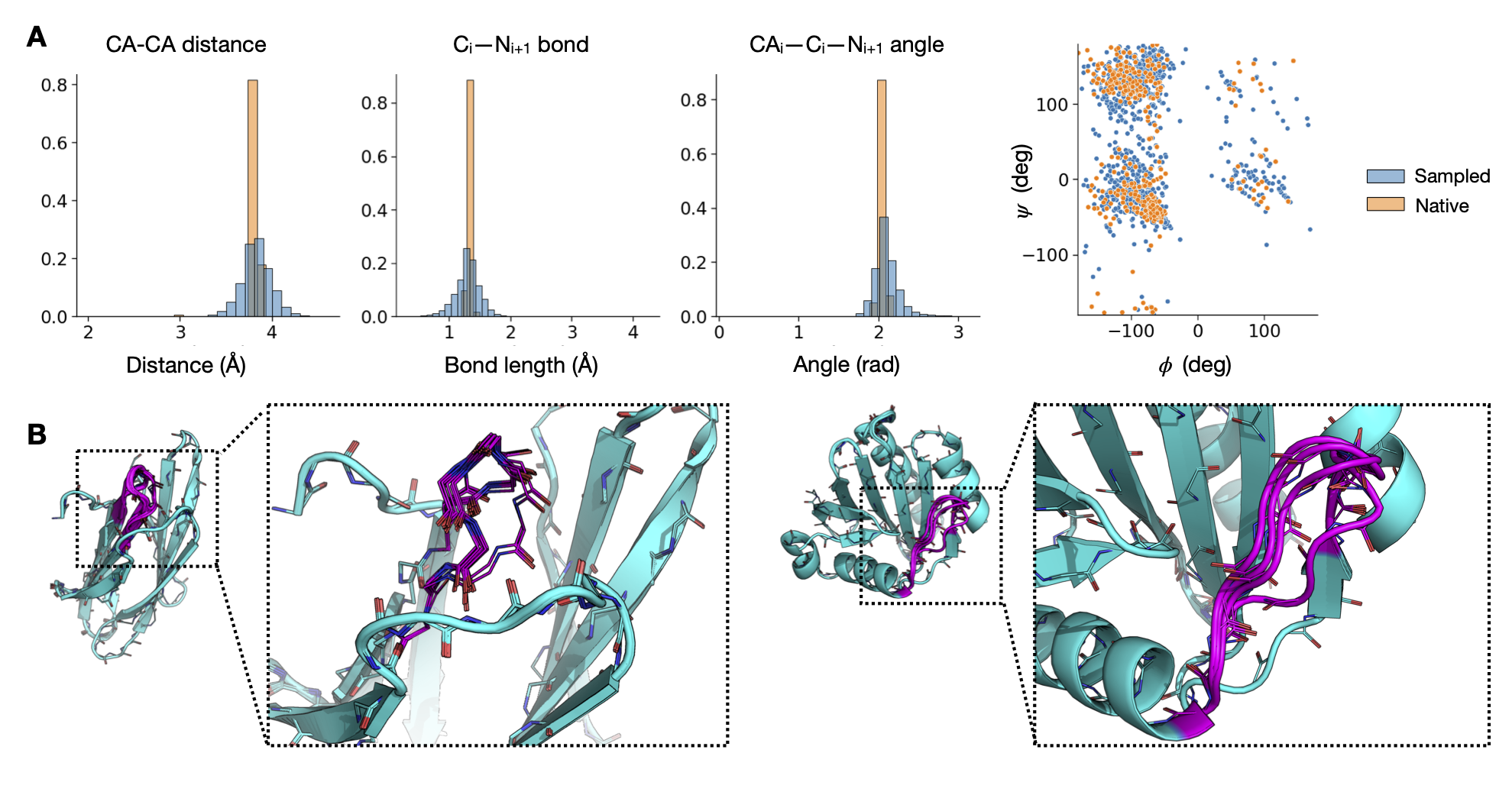
Structure Inpainting
Inpainting α-helices and β-sheets.
Capturing multiple discrete modes of loop geometry.
End to End Immunoglobulin (Ig) Loop Backbone and Sequence Generation
Designing Ig loop backbones and sequence jointly, followed by rotamer packing.
Citation
@misc{anand2022protein,
title={Protein Structure and Sequence Generation with Equivariant Denoising Diffusion Probabilistic Models},
author={Namrata Anand and Tudor Achim},
year={2022},
eprint={2205.15019},
archivePrefix={arXiv},
primaryClass={q-bio.QM}
}